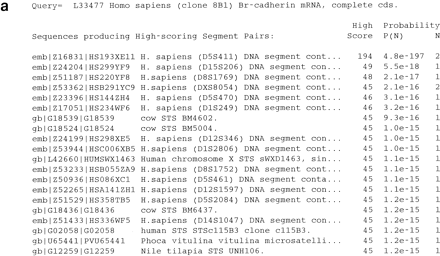

BLAST search with a microsatellite-containing query sequence. The BLASTN program was used to search the dbSTS database with the sequence corresponding to GenBank entry L33477 (Br-cadherin) as the query sequence using a match score (M parameter) of 1 and a mismatch score (N parameter) of −2. The query sequence was not filtered for low-complexity sequences. (a) The first 20 sequences listed on the resulting “hit list” are shown, sorted by statistical significance. Altogether, >8000 sequence matches were found (by default, only the first 500 are shown, but the complete list can be obtained by setting the V parameter to a very large number). The best match was observed against the sequence corresponding to GenBank entryZ16831, which contains the Généthon marker D5S411. When low-complexity filtering is used, the problem is reduced dramatically, but 10 false positives remain so manual inspection of the results is still required. (b) The sequence alignment generated by BLAST for the sequence corresponding to GenBank entry Z24204, which was the second-best hit reported, contains the Généthon marker D15S206. The alignment includes only the (CA)nmicrosatellite repeats.











