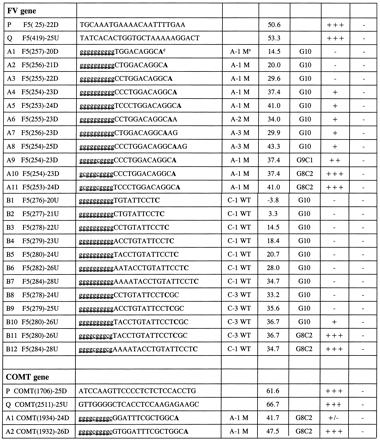
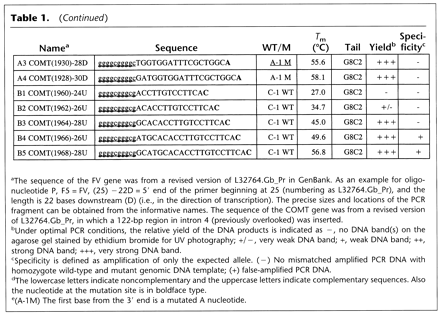
Selected Primers for the Initial Optimization of FV and COMT Genes
| Name | Sequence | WT/M | T m (°C) | Tail | Yield | Specificity | |
|
|||||||
|
|||||||
-
↵The sequence of the FV gene was from a revised version of L32764.Gb_Pr in GenBank. As an example for oligonucleotide P, F5 = FV, (25) −22D = 5′ end of the primer beginning at 25 (numbering as L32764.Gb_Pr), and the length is 22 bases downstream (D) (i.e., in the direction of transcription). The precise sizes and locations of the PCR fragment can be obtained from the informative names. The sequence of the COMT gene was from a revised version of L32764.Gb_Pr, in which a 122-bp region in intron 4 (previously overlooked) was inserted.
-
↵Under optimal PCR conditions, the relative yield of the DNA products is indicated as −, no DNA band(s) on the agarose gel stained by ethidium bromide for UV photography; +/−, very weak DNA band; +, weak DNA band; ++, strong DNA band; +++, very strong DNA band.
-
↵Specificity is defined as amplification of only the expected allele. (−) No mismatched amplified PCR DNA with homozygote wild-type and mutant genomic DNA template; (+) false-amplified PCR DNA.
-
The lowercase letters indicate noncomplementary and the uppercase letters indicate complementary sequences. Also the nucleotide at the mutation site is in boldface type.
-
(A-1M) The first base from the 3′ end is a mutated A nucleotide.











