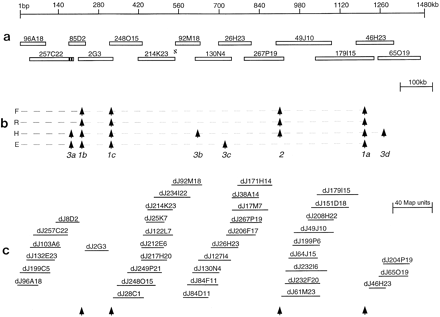
Comparative fingerprinting data of 49 clones covering a 1400-kb interval sequence that was determined from 14 clones comprising a minimum tiling path and a PCR product between 214K23 and 92M18 (hatched). (a) Finished sequence data were used to define the overlaps between clones and absolute length of each of the clones (except 257C22, where data were incomplete). (b) Vertical arrows denote overlaps that were not detected by each method. (F) Fluorescence, (R) radioactive; (H) HindIII, and (E)EcoRI. As expected, there is variation based on recognition sites between the overlaps detected using HindIII (3band 3d) and those established with EcoRI (3c). Details of the analysis of undetected overlaps are as follows: (Overlap 1a, 62 kb) Nine HindIII and 11EcoRI fragments are present in this overlap on the basis of DNA sequence. Statistical probability on the basis of these bands shared between 179I15 and 46H23 is 1 × 10−4 (−4) fluorescence, −1 radioactive, −4 HindIII, and −5EcoRI (whose probability cutoffs for all matches are −4, −5, −7, and −7, respectively). (Overlap 1b, 25 kb) Clone 2G3 is unattached. Three HindIII or six EcoRI fragments shared with clone 85D2 are insufficient to score the overlap. (Overlap 1c, 11 kb) Clone 2G3 is unattached. FourHindIII or three EcoRI fragments shared with clone 248O15 are insufficient to detect the overlap. (Overlap 2, 24 kb) Overlap between 267P19 and 49J10 was not found by any of theHindIII methods. Six HindIII fragments are not sufficient to detect overlap. Arrows 3a–3d denote where only the agarose methods failed to find overlap. (Overlap 3a) Overlap between 257C22 and 85D2 could not be determined because of incomplete overlap data and therefore was not analyzed. (Overlap3b, 18 kb) Four HindIII fragments shared between 130N4 and 92M18 are not sufficient to detect overlap. (Overlap3c, 44 kb) Six EcoRI fragments shared between 130N4 and 267P19 are not sufficient to detect overlap. (Overlap 3d,55 kb) 16 HindIII fragments between 46H23 and 65O19 should have been significant. The lack of overlap may be attributed to the migration of similar-sized fragments in the same location of the agarose gel and therefore not be representative of the cloned insert. (c) Although all but 1 of the 49 clones (2G3) fell into contigs, only 4 of the contigs that lie within the sequence interval are represented. Arrows denote where the overlaps between clones were not found.











