WebWise: The Washington University Genome Sequencing Center’s Web Site
The first web site review in the WebWise series is an overview and navigation guide for Washington University Genome Sequencing Center’s (GSC) web site (http://genome.wustl.edu/gsc/gschmpg.html). This sequencing center has established a high-throughput DNA sequencing facility and has committed the resources required to establish and maintain a World Wide Web site. Those who have been involved in establishing and maintaining a web site will understand that it is a large effort and requires a continuing commitment. Thus, while acknowledging the wealth of data available at these web sites, this review will also acknowledge when the data presentation is inconsistent or incomplete. It is important to note, however, that web sites continue to evolve and often undergo major revision. Therefore, each review reflects the status of the web site prior to publication of each issue. Although each report provides some comparison across a set of standard features found at many sites, each survey will also highlight any special features available at the web site. Reviews will emphasize the main features of these web sites and provide an overview of the general organization, informational resources, map and sequence data presentation and availability, search services, and availability of additional software tools. The main intent of this series is to provide a guide to the location, quantity, and quality of the data to facilitate your use of each web site. Finally, it may be useful to have each web site open on your computer as you read each article.
Washington University GSC
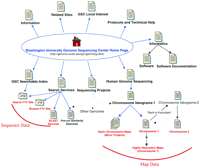
Washington University’s GSC web site provides a good illustration of the utility of a web guide. This web site is large and complex, includes a variety of informational content, and presents initial navigation challenges. GSC provides a lot of data on their web site; at the time of this writing, they report having finished >19 million bases of human DNA sequence. Chromosomes 2 and 7 are their main sequencing focus, but they are also involved in sequencing regions of chromosomes 3, 5, 8, 12, 13, 16, 22, and X. The site map depicted in Figure 1 illustrates the overall organization of this web site, and the main features of the web site are highlighted in Table 1. Note that many internal, duplicate, or minor links have been omitted from the diagram. From the Home page, the user is presented with several possible directions to explore, which can be categorized as (1) general information, (2) data, and (3) tools.
Washington University GSC site map. The main links to pages discussed in the text are illustrated here. Links on the top of the Home Page are to general informational resources; links located on the bottom portion are to the data or additional tools.
Features of the Human Genome Project Web Sites
General Information
The links located on the top of the diagram (Information, Related Sites, GSC Local Interest, and Protocols and Technical Help) all lead to general information related to the genome sequencing effort. Some of the information presented on the Information web pages is of specific interest, such as travel directions; but the GSC Manual (http://genome.wustl.edu/gsc/manual/protocols/Cover95.html—located from the Protocols and Technical Help page) provides detailed sequencing protocols that might be of use to laboratories beginning a large sequencing project or engaged in experimental troubleshooting. An additional user’s manual on Script and Software is available from the GSC manual page but not very well advertised elsewhere. The Script/Software Table of Contents link calls up a list of sequencing software tools such as those used in preprocessing (abi2phred and ASP), “mid”-processing (Consed, Staden Package), and finishing (Hawk, Squawk, Exp-view, and dotter), which are documented in the Scripts/Software manual. This is a useful resource for those who are not yet experts in the large-scale sequencing field and provides documentation on the purpose, use, and possible errors encountered with various sequencing-related software tools. Unfortunately, the description of the various programs is inconsistent, some of the write-ups appear to be primarily targeted toward in-house use, and no mention is made of public availability of most programs discussed. The web site does clearly indicate that the Scripts/Software manual is still under development, which likely explains both the above observation and the lack of a link from the Informatics web page.
Data
The data actually consist of both maps and actual DNA sequence data. If, for example, you are interested in downloading unfinished sequence and/or following the progression of sequencing on a particular chromosome near “your favorite gene,” you will probably have to begin by examining the chromosome maps provided for that region to ascertain the clone names and their sequencing status. Following the Home page link to Human Genome Sequencing (http://genome.wustl.edu/gsc/Web_pages/HGseq.html#Top) brings you to a page reviewing those chromosomes targeted for sequencing. As you scroll down the page you see a series of chromosome ideograms with, for some chromosomes, a general summary about the size of the target, clone origins, references, and links to collaborators. To access the individual chromosome maps of those regions targeted for sequencing, follow the links provided under the chromosome ideograms. Two different styles are used for the chromosome maps; dynamic image map diagrams are available for the major sequencing effort of chromosome 7, and static (nonclickable) map diagrams are provided for the “minor” chromosome efforts (3, 5, 8, 12, 13, 16 22, X). The chromosome 2 sequencing effort has not matured yet; maps are not available, and the small amount of sequence data generated are not accessible from this page (see below for additional information).
The chromosome 7 project represents ∼90% of the human sequencing effort at the GSC. These maps are kept up to date, as the dynamic image mapped displays are automatically generated and updated daily. Because most of chromosome 7 is targeted for sequencing, the chromosome is successively divided into regions and targets that you must click on to narrow down your region of interest. After several clicks you are presented with an unordered list of contigs covering the targeted region. Clicking on a contig will call up a static map displaying the clone tiling path and, when available, a color-coded status bar. These pages were updated while this paper was being written, and a considerable number of new contig maps are now available. Although an impressive amount of data are represented by the chromosome 7 maps, it is somewhat challenging to get to the data for any given targeted region until you have determined which target and contig(s) you are interested in. As it stands now, one must successively click on the unordered contig links to find one for which some data are available. The lack of any indication of which targets and contigs have available DNA sequence data makes it extremely difficult to determine the overall progress of the chromosome 7 sequencing project from these maps. From a user’s perspective, it would be very useful if the maps included a general status indicator (indicating some sequence is available on the FTP site) for each successively higher-resolution map view. It should be noted that there is preliminary, finished, and archived chromosome 7 sequence data available on the FTP site; yet it is difficult to determine which general regions of chromosome 7 these sequences correlate to without manually “walking” through all of the contig links for each target—a time-consuming and tedious job, to say the least.
The chromosome 2 sequencing effort has only recently been initiated, and its scope and status are similarly obscure. The main ideogram display does not provide a link to any chromosome 2 data; however, the top of the first chromosome 7 page includes a Back to Karyotype link, which brings you to a second ideogram view that does include a chromosome 2 link. A chromosome 2 map is not yet available, but a clone of uncertain status is listed and is hot linked to the DNA sequence.
The map diagrams displayed for the targeted regions of chromosomes 3, 5, 8, 12, 13, 16, 22, and X impart significant information by including an indication of the sequencing group identity, the clone order and name, and the status of each clone. These maps are not image mapped, but rather are static (nonclickable) images, which are updated every 2 weeks or as needed. The color-coded rectangles on the left side of each diagram reflect the identity of the in-house group working on that clone, whereas the color-coded regions on the right side of the diagram indicate the actual status of the DNA sequence for a given clone. Although these maps contain a lot of useful information, interpreting diagrams can be somewhat confusing, especially given the reuse of the same color key for two different things. In addition, there is some inconsistency in the diagrams, which hampers interpretation further. For instance, some of the more complex diagrams (see chromosomes 22 and Xq23) include the group key and clone name on the left side, an additional color-coded line in the center, and the color-coded clone sequence status rectangles on the far right. It is not clear exactly what the color-coded line in the center represents. These pages would benefit from the addition of some explanatory text describing the various features of the diagrams.
Traversing the connection between the map data and the DNA sequence data is not immediately intuitive. Unfortunately, the most intuitive method, that of providing an image map in which the clones are hot linked directly to the DNA sequence, is not provided here. You can find the DNA sequence data by following the Home page link to the Search Services page. From that page you can choose to browse the FTP site (ftp://genome.wustl.edu/pub/gsc1/sequence/st.louis/human/) or search it using the GSC Searchable Index (see below). If you browse the ftp site, the sequence is initially divided into a series of folders by organism. The human sequence folder is further divided into three folders containing sequences that are unfinished, finished, and submitted to GenBank. Data are updated nightly, and as sequence status is updated, the sequence file may move to a new folder on the FTP site. Once you have determined the clone name, by looking at the maps, it is an easy matter to keep downloading that sequence file from the ftp site. Although the data are publicly available, a data release statement is located on the FTP site (ftp://genome.wustl.edu/pub/gsc1/sequence/README).
Tools
In addition to providing an abundance of map and sequence data, this web site provides some additional features, including a web and ftp search tool, the means to carry out BLAST searches against the GSC databases, archived prerun similarity searches, and a description of the software tools used at the GSC. These features further enhance the overall utility of this web site by making it that much easier to find the data, compare your sequences to their data, and perhaps identify meaningful sequence homologies.
Washington University’s GSC has provided a convenient search tool, the GSC Searchable Index (accessible from the Home page, the Search Services page, or the Chromosome Ideogram page), to locate sequences on the FTP site. You can also use this tool to search the web site, although the FTP search capability seems more useful. The GSC Searchable Index is easy to use, responds quickly, and provides a link to the DNA sequence in FASTA format. In some cases, a link is also supplied to a prerun similarity search document relating some sequence annotation including statistical information about the sequence such as the %GC content, frequency of repetitive sequences, and any identified homologies. Although the prerun similarity searches do contain some generally useful information, there is no indication that the homology searches are repeated as the public databases grow, so the available homology information must be taken as a preliminary indication only. The GSC Searchable Index approach worked very well when tested with clone names from the chromosome 22 and X maps; however, searching for several chromosome 7 clone names (obtained by looking at the chromosome 7 contig maps) did not give any results. A second search engine, called DACE, is under development and can be found by following the Search our Databases link from the Search Services page. This search engine searches the ACEDB databases; although it is not fully implemented yet and is limited to Caenorhabditis elegans data, it appears to be a good approach to providing searchable map data. As DACE was not yet able to search the human genome databases at the time of this writing, it is not reviewed in detail here.
The Human BLAST server is an important tool for a sequencing center web site to provide. This feature enables users to submit their own sequence, in FASTA format, and perform a BLASTN, TBLASTN, or TBLASTX query against the GSC sequence database. This search service was tested with a sequence that was selected because it should return a result, in this case a mouse BRCA2 cDNA sequence. Although fastA is stipulated as the format to use, the initial line of a fastA file (>text) did generate some error warnings, but these did not impact the result returned to the screen. The result is returned to the web browser screen within, on average, 3 min during daytime usage. The user may alternatively choose to have the result returned to an e-mail address. The output consists of a list of homologous clones, scores, and alignments; however, these data are not linked to either the maps or the DNA sequence data. Nevertheless, this tool is very useful as it enables users to identify homologous sequences located in the GSC databases; if one is working with a cDNA or otherwise short DNA sequence, identifying a region of homologous genomic DNA sequence can have obvious benefits.
Following the Home Page Informatics link leads you to a list of all programs used at the GSC. Detailed software documentation, including instructions for obtaining a copy of the program, is available for three packages. (1) The ACEDB [A C. elegans Database,Eeckman and Durbin (1995)] program and documentation is available from several anonymous FTP servers (seehttp://probe.nalusda.gov:8000/acedocs/whereacedb.html). (2) Documentation on the ContigC mapping program is available on the web site as well as instructions for obtaining it from an anonymous FTP site (ftp.sanger.ac.uk/pub/contigc). (3) The Staden Package, a compilation of sequence assembly and analysis programs, documentation is also available on the web site; general instructions on obtaining this package, including a link to the required agreement form, are provided (see http://genome.wustl.edu/gsc/new/staden/blurb.html). A complete list of all the scripts used at the GSC is also available. Although this is a nice feature, it is not clear whether this software is available by FTP, or by any other route. Directories bearing an obvious software-related name were not noticed on the FTP site.
Conclusions
The Washington University GSC web site presents a wealth of both map and sequence data to the user. The main features available at this web site are highlighted in Table 1. The sequence data are updated nightly, and map data appear to be updated more sporadically. Most of the main pages include a navigation link on the top of the page, which greatly increases general ease of use for those pages. However, these internal links are not supplied for the more internal pages, which makes it more difficult to navigate from, for instance, a higher resolution map to the GSC Searchable Index. Although the data available at this web site are a valuable resource to the research community, the map data in particular do not utilize a uniform style and can be confusing to interpret. The lack of a uniform style makes it more difficult to extract the relevant information, and the lack of a comprehensive “big picture” for chromosome 7 makes it very difficult indeed to identify which sequence files pertain to particular regions. The enhanced features available at this site, including the GSC Searchable Index and the Human BLAST server, are quite useful. In general, the correspondence between the map data or BLAST results and the sequence data would be enhanced by the inclusion of links from the clone names to the sequence file.
Footnotes
-
↵1 E-MAIL ; FAX (301) 435-2433.
-
Next month: The Sanger Center
- Cold Spring Harbor Laboratory Press