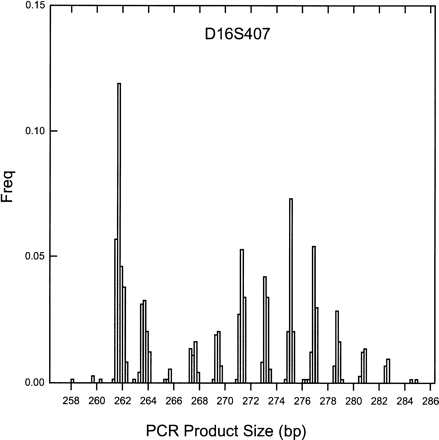
Figure 3.
Frequency histogram of illustrative marker which has nonstandard repeat lengths between adjacent alleles. In this dinucleotide marker, the expected difference between alleles is 2 bp, yet the average spacing is 1.90 bp. The automated binning procedure accurately assigns all bins only when allowing for this discrepancy of repeat length, referred to as allelic drift.











