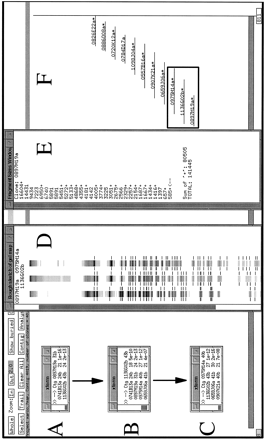
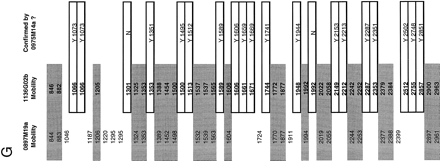
A specific example of contig construction, on human chromosome 7, in FPC. Contigs are constructed as described in Methods (computer analysis and contig construction). Windows A, B, andC show the results of searching individual clones against FPC. The PAC clones 0897M19a, 1136G02b, and 0975M14a, used here to query the FPC database, correspond to the theoretical “clone 1,” “clone 2,” and “clone 3” described in Methods (computer analysis and contig construction). Window D shows the FPC fingerprint viewing tool displaying the fingerprints of these three clones as a representation of the original agarose gel fingerprint data output from the program Image. Note, however, that the lanes displayed are substantially altered versions of the original agarose gel image collected on the Molecular Dynamics FluorImager. The band positions have been normalized to the band positions of the marker DNA and only a one pixel-wide “slice” of the original 17 pixel-wide agarose gel image can be displayed. Manually verified band positions are indicated by the hashmarks flanking the lanes. Only these bands are used by FPC in database comparisons. Other band-like entities present in the lanes correspond to computer artifacts generated during the original gel image modification for display in FPC, to fluorescent foreign particles embedded in the agarose detected by the FluorImager during data acquisition, and rarely to legitimate bands that were not identified during the manual band calling routine conducted in Image. Examination of the original agarose gel images, retained in hard copy and electronic format, allows resolution of these alternate possibilities. Window E shows the restriction fragment sizes (in base pairs) for the clone 0897M19a. The + symbols to the right of the fragment sizes indicate fragments that have been user-selected by clicking with a mouse on the hashmarks in the FPC fingerprint viewer (D). Clicking a second time on the hashmark deselects the fragment. The sum of the sizes of the selected fragments is given at the bottom of window E, as is the total estimated size of the clone. The total size is calculated by summing the sizes of all the restriction fragments. This latter feature is a non-FPC function added at Washington University. Window F is an FPC display of a manually generated diagram of the contig after analysis is complete. The asterisks beside some clones indicate that redundant clones are contained entirely within these clones. Redundant clones are referred to as “buried” clones and are not visible in this view. A total of 40 clones have been incorporated into this contig. The clones considered specifically in this example are indicated with a box. Not shown are the additional search results used to construct this contig.G provides an example of the raw data used by FPC in performing the calculation of the Sulston score. Shown are the normalized relative mobilities (in tenths of a millimeter) for all of the detected restriction fragments for the clones 0897M19a and 1136G02b. The shaded values are considered common between these two clones. The values contained within open boxes indicate fragments unique to 1136G02b, all of which (except two fragments that are presumed to be the anomalous vector-insert junction fragments found in PACs digested with HindIII) are “confirmed” (Methods) by their prescence in the clone 0975M14a. Only the confirming fragments for 0975M14a are shown. These are indicated with a Y (for Yes). Details of FPC search results: The result of searching a PAC clone 0897M19a against the FPC database is given in Figure 2A. Shown in the window labeled xterm are the Sulston scores (see Methods) for the PAC clones 0741B13a and 1136G02b. The smaller score, indicating relatively more extensive overlap with 0897M19a (Methods), is exhibited by 0741B13a. Manual comparison of the fingerprints (not shown) of these two clones revealed that 0741B13a (24 restriction fragments) contains only two novel restriction fragments, which may correspond to the PAC vector-insert junction fragments. Manual comparison (Figure 2D) of the 1136G02b fingerprint (43 restriction fragments) with that of 0897M19a (31 restriction fragments) revealed that 20 of these fragments were unique to 1136G02b. 1136G02b was then used to query FPC. The results of this search are shown in Figure 2B. Strong matches to the previously mentioned clones are indicated, as are matches to the clones 0975M14a (40 restriction fragments) and 0659J06a (41 restriction fragments). Manual comparison of the 0975M14a fingerprint to that of 1136G02b (Figure 2D) reveals that the restriction fragments identified as unique to 1136G02b in the 1136G02b–0897M19a comparison are present in the fingerprint of 0975M14a (see Fig. 2D,G). These fragments are thus said to have been “confirmed” (Methods) and 0975M14a can be incorporated into the nascent contig.











