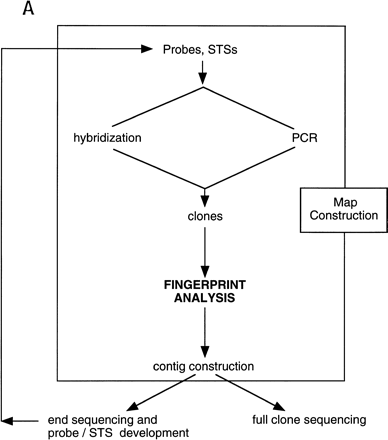
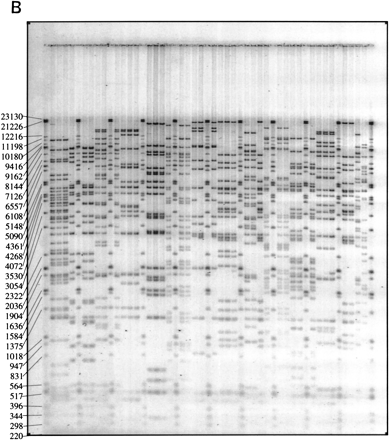
Schematic illustration of the role played by fingerprinting in the construction of sequence-ready contigs. (A) For construction of “local” sequence-ready maps, STSs, and probes specific for a small region, typically 1–2 Mb, are used to identify clones, either by hybridization or PCR methods. The fingerprinting produces higher resolution clone-specific information and facilitates contig construction and selection of clones for sequencing. Intercontig gaps are closed by chromosomal walks using probes developed from end sequences. Clones identified during walks are fingerprinted to determine their relationship to the contigs, permitting recognition of clones spanning intercontig gaps. (B) A typical agarose-mapping gel showing human PACs digested with HindIII. Clones are present in triplicate to verify stability during propagation and to control for the possibility of cross-contaminated glycerol stocks in the intial 384-well format. DNA size standards, which are a mixture of three commercially available markers (see Methods), are present every fifth lane. The sizes, in base pairs, of the marker fragments are indicated.











