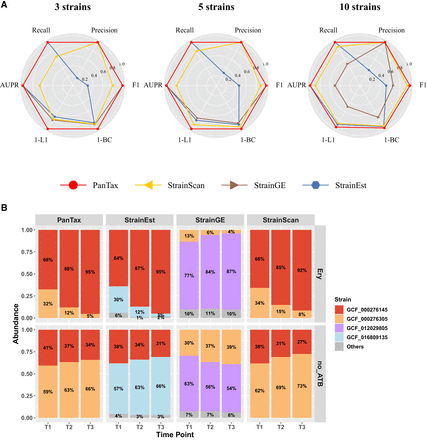
Benchmarking results of single species strain-level taxonomic profiling. (A) Simulated data sets (S. epidermidis strain mixtures: three strains, five strains, and 10 strains). (AUPR) Area under the precision-recall curve. To visualize all metrics consistently (i.e., with higher values indicating better performance), we present the 1-L1 distance and 1-BC distance. (B) Real data sets: two-cultured S. epidermidis strain mixtures. The x -axis represents time points, and the y -axis shows the relative taxon abundance of each identified strain. The two strains (marked in red and yellow) were mixed in equal proportions (1:1) and cultured under two conditions: one with erythromycin treatment (Ery) and one without antibiotics (no_ATB). The upper and lower panels display the strain abundances predicted by four tools under Ery and no_ATB, respectively.











