Table 4.
Accuracy of classification methods
| Model | Criterion | All | EUR | EAS | AMR | SAS | AFR | OCE | WAS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TR | TS | TR | TS | TR | TS | TR | TS | TR | TS | TR | TS | TR | TS | TR | TS | ||
| PCA | 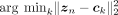 |
78.6 | 74.1 | 64.9 | 66.3 | 71.1 | 74.3 | 87.2 | 77.8 | 58.5 | 57.2 | 97.5 | 93.4 | 95.4 | 76.6 | 75.6 | 73.4 |
| VAE | 93.2 | 85.7 | 81.7 | 78.6 | 96.9 | 96.3 | 99.5 | 92.1 | 81.6 | 78.5 | 99.3 | 96.8 | 99.4 | 71.7 | 94.0 | 86.1 | |
| C-VAE | 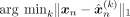 |
93.4 | 78.0 | 84.4 | 70.6 | 96.4 | 92.1 | 99.9 | 92.4 | 84.4 | 76.4 | 99.9 | 96.8 | 100 | 62.8 | 88.5 | 54.7 |
| arg maxk p(Y = k|xn, θ) |
97.5 | 87.1 | 96.4 | 87.1 | 98.5 | 95.5 | 100 | 97.4 | 90.0 | 81.2 | 99.4 | 94.9 | 100 | 79.8 | 98.1 | 73.5 | |
| Y-VAE | 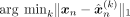 |
98.9 | 83.2 | 96.9 | 81.5 | 99.6 | 96.2 | 99.9 | 87.2 | 98.5 | 90.1 | 100 | 98.4 | 100 | 68.6 | 97.5 | 59.9 |
| arg maxk p(Y = k|xn, θ) |
99.1 | 85.2 | 97.6 | 84.3 | 99.7 | 96.6 | 100 | 90.9 | 98.2 | 88.5 | 100 | 98.2 | 100 | 72.7 | 98.3 | 65.0 | |
-
TR refers to accuracy computed on training data and TS on test data, accordingly. The values represent the accuracy in %. Note that regular VAE, C-VAE, and Y-VAE have 10,371,760, 10,378,928, and 72,602,320 parameters, respectively. Bold values indicate the highest accuracy (best performance) on test samples across the compared models and criteria.











