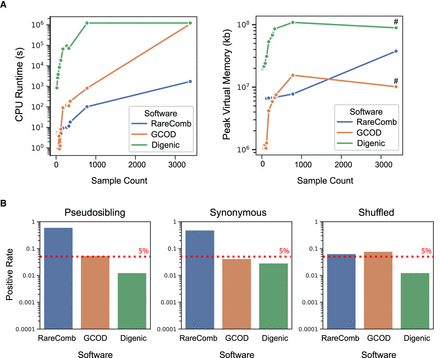
GCOD efficiently evaluates oligogenic gene sets while limiting type-I errors. (A) CPU runtime (left) and peak RAM usage (right) of GCOD and two other cohort-based oligogenic discovery software tools. Analyses were completed for the full PCGC cohort as well as smaller subdiagnoses of CHD, providing a range of sample sizes (x-axes). Across a range of sample sizes, GCOD's runtime is slower than RareComb's but faster than Digenic's, becoming equivalent to Digenic for the full cohort of 3377 trios (left). Peak memory usage is lower for GCOD compared with the other methods at most sample sizes (right). We note that peak memory, unlike CPU time, does not sum across multiple parallel threads; this explains the drop for peak memory in GCOD and digenic at higher cohort sizes, which were parallelized (indicated by # for parallelized runs). (B) Type-I error rate of the three methods, using pseudosibling variants with CADD scores in the top 0.5% exome-wide (left), proband synonymous variants (center), and variants with shuffled family IDs (right) as controls. GCOD controls the false-positive rate at ∼5% (dashed line), whereas Digenic is relatively conservative and RareComb is anticonservative.











