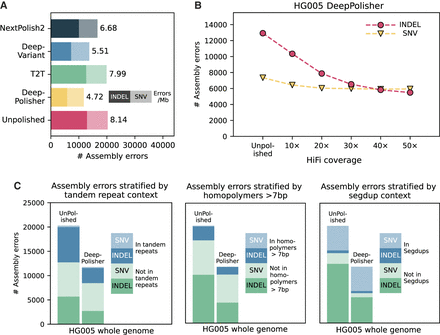
Comparison of DeepPolisher and alternate polishing methods against GIAB v4.2.1 benchmark for HG005. (A) For each polishing method, GIAB v4.2.1 variant-calling (assembly) errors are separated by indels (darker shade) and single-nucleotide variants (SNVs; lighter shade), with the number of errors per megabase to the right of each bar. (B) Total GIAB variant-calling (assembly) errors for different HiFi read coverages, with indel errors represented in pink circles and SNV errors in yellow triangles. (C) Total GIAB variant-calling (assembly) errors stratified by presence in tandem repeats (left), homopolymers >7 bp (middle), and segmental duplications (segdups; right), with SNV errors in lighter shades and indel errors in darker shades.











