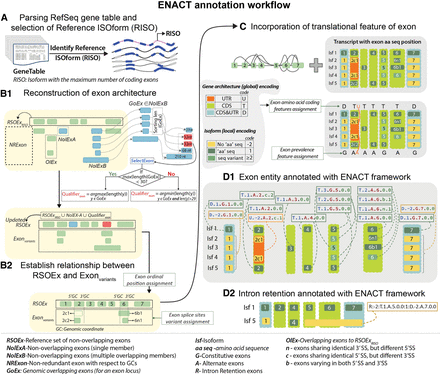
Overview of ENACT annotation workflow. Figure illustrates the main steps of the ENACT algorithm (A–D). (A) Curated models of protein-coding transcripts or isoforms with their genomic and coding genomic coordinates are collected in gene table format from NCBI RefSeq (blue and gray colors represent coding and noncoding exons, respectively). A reference isoform (RISO) is selected based on the maximum number of coding exons (see Methods). (B1) The initial set of reference exons (RSOExRISO) consists of exons from RISO, which are compared with nonredundant exons (NRExon) from other isoforms using genomic coordinates. This yields a set of overlapping (OlEx) and nonoverlapping exons (NolEx). NolEx entities without varying genomic coordinates are categorized as NolExA and are added to RSOEx, whereas other NolEx entities (NolExB) are further processed to yield subsets that show genomic coordinate overlaps for a locus, referred to as GoEx. One such GoEx entity is illustrated in B1 (penultimate exon). GoEx sets are iterated to select representative exons (Qualifierexon) for the RSOEx, based on exon length using “selectExon” procedure (see Box 1). The updated RSOEx includes NolExA and Qualifierexon from GoEx subgroups. Similarly, the Exonvariants set includes initial OlEx and exons from GoEx subgroups, excluding their Qualifierexons. (B2) Exonvariants entities are compared with RSOEx to define alternate splice-site variants: 5′, 3′, and 5′ with 3′ are characterized as “n,” “c,” and “b,” respectively. (C) The RSOEx and their associated splice variants from the Exonvariants set are sorted based on genomic coordinates and assigned ordinal positions (from one to the number of exonic loci in RSOEx). After establishing updated RSOEx and others as splice-site variants, exon translation attributes are assigned considering coding genomic coordinates. Further, prevalence attributes are noted based on locus prevalence across isoforms: (G) constitutive, (A) alternate. This procedure annotates each exon in RSOEx and Exonvariants with its relative position, translational (protein-coding potential) feature, occurrence, and splice-site variations. (D1) Exon attributes are combined to construct a six-character alphanumeric notation defined as an exon unique identifier (EUID). For the representative example, EUIDs for each exon are shown. (D2) The intron retention instances are identified in step B and annotated with intron retention (IR) codes.











