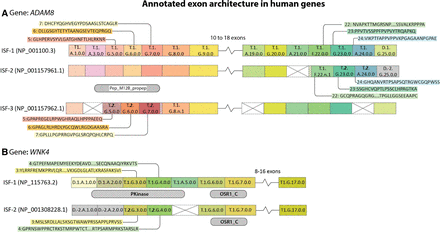
ENACT annotation of ADAM8 and WNK4 isoforms and exons. (A) In ADAM8 isoforms, ISF-2 undergoes a reading frame shift in the C-terminal region, involving Block-I local scope variation in exons 23 and 24. A similar reading frame shift occurs in ISF-2 in the N-terminal region, resulting in a loss of the “Pep_M12B_propep” domain. This change in reading frame is noted through Block-I local scope variation in exons 5, 6, and 7. (B) In WNK4, ISF-2 lacks a “Pkinase” domain in the N-terminal region and undergoes reading frame shift in exons 3 and 4, as indicated by their Block-I local scope values. The NCBI gene identifier is shown for each isoform, and exons are represented as colored rectangle boxes with their respective EUID. The skipped exon is shown with a crossed empty rectangle box. The small extension of exon rectangle boxes with crisscross-filled lines represents extended exon boundaries owing to alternate splice sites (5′/3′). Exons not showing any variation are left unlabeled. The break shows that exons intervening in the region do not change in the isoforms. The jagged edge of a rectangle represents alternate 5′ or 3′ splice site. The isoforms sharing Pfam domains for a region are shown below the exon layout. Changes in Block-I local scope are complemented with sequence specifications.











