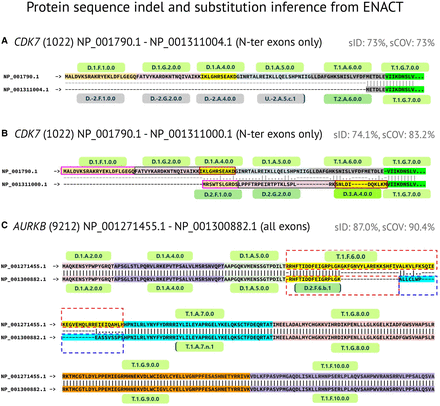
Features of ENACT EUIDs in inferring protein indels and substitutions. Protein sequence alignment of isoforms, overlayed with exon entity, for three genes are shown to highlight indels/substitutions. (A) N-terminal indel introduced by “dual” exons in CDK7 gene, in which their noncoding instances (exons 1, 2, and 4, and a splice variant of 5) are missed in ISF: NP_001311004.1 sequence respective to ISF: NP_001790.1, resulting in isoforms having 73% of sequence identity and coverage. (B) N-terminal substitution (CDK7), in which dual exons 1, 2, and 4 contribute different amino acid sequences in ISF: NP_001311000.1 in comparison to ISF: NP_001790.1 owing to alternative translation initiation site (also depicted in Fig. 3A). The altered reading frame in ISF: NP_001311000.1 is rescued after skipping of the fifth and sixth exons. (C) EUID-enabled substitution inference is illustrated in the middle of the ISF: NP_001300882.1 sequence compared with ISF: NP_001271455.1 of the AURKB gene. Ordinal exon position 6 shows splice-site alteration (recorded by Block-III subtype “b,” occurrence value: 1) in ISF: NP_001300882.1. In ISF: NP_001300882.1, this instance contributed a different sequence, which aligns with the N-terminal of the reference instance. Another event record in ISF: NP_001300882.1 is noteworthy, in which exon 7 undergoes 5′ splice-site alteration (recorded by Block-III, subtype “n,” occurrence value: 1) and contributes an extended amino acid sequence. Exon 6 and exon 7 events are represented in red and blue dashed boxes. Protein sequence alignment was performed using the Needleman–Wunsch algorithm for isoform pairs. “sID” and “sCOV” refer to sequence identity and coverage, respectively. The amino acid sequences are highlighted in distinct colors to demarcate their respective exon with their corresponding EUIDs, which are listed above for the top isoform and below for the other isoform in the alignment. No EUIDs are specified for skipped exons.











