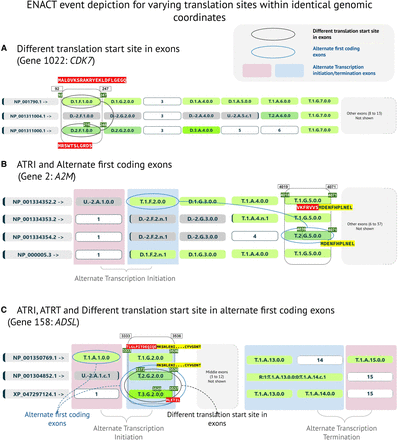
ENACT depiction of translation start/end site and transcription start/end site variations. (A) Sequence changes in the first exon of human CDK7 gene are shown for ISF: NP_001790.1 and ISF: NP_001311000.1 owing to a change in translation initiation site as indicated by different 5′ coding genomic coordinates (black oval). This variation is captured in Block-I local scope within the EUID, as it has different values for exon 1 in two isoforms. The amino acid sequences encoded by each exon entity are also shown. (B) The utilization of alternate first coding exons is shown for human A2M gene isoforms involving exon 2 and exon 5 (blue oval). This indicates alternate translation initiation sites in isoforms through different exons. The sequence variations in exon 5 are shown and are captured in Block-I's local scope. This variation leads to considerable N-terminal sequence truncation in ISF: NP_001334354.2. (C) This panel illustrates variations in both alternate first exons (blue ovals) and alternate translation sites (black oval) in three isoforms of human ADSL gene. Exons 1 (ISF: NP_001350769.1) and 2 (ISF: NP_001304852.1 and XP_047297124.1) in the coding subspace are alternate first coding exons in respective isoforms. Exon 2 exhibits two translation initiation sites in ISF: NP_001304852.1 and XP_047297124.1, depicted using Block-I local scope values of two and three. Exon alignment obtained from ENACTdb represents alternate translation initiation sites. Genomic boundary coordinates for exons of interest across isoforms are indicated with values at the top of the rectangular box. Coding genomic coordinates are mentioned in the exon block with an olive-colored text background. Vertical pink and light blue backgrounds for exon positions highlight the alternate first or last exons related to alternate transcription in different isoforms.











