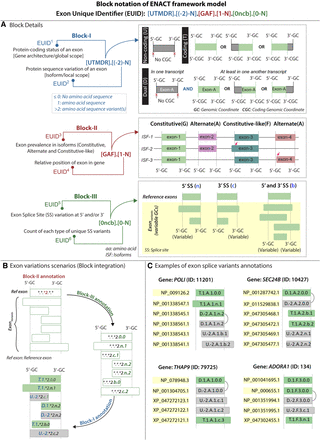
Overview of ENACT exon nomenclature. (A) Blocks of EUID represent several exon feature variations. Block-I global scope defines translational features of exons with unchanged genomic coordinates (GCs) by considering their coding genomic coordinates (CGCs). Exons are depicted as having coding, noncoding, and dual scope, where the “dual” is assigned to exons that take part in both coding and noncoding states in different isoforms. Block-I local scope depicts amino acid sequence contributions, where values of greater than one inform sequence variation from an exon and values of zero or less inform no amino acid sequence. The latter category involves (1) UTR exons that have no CGC and no amino acid assigned, depicted as “−2,” (2) exons having only 1 nt in CGC, which cannot contribute amino acid independently, depicted by local scope “0” and often global scope “M,” and (3) exons having >1 nt in CGC but no assigned amino acid, albeit rare in occurrence depicted by local scope value “−1.” Block-II represents exon occurrence as “constitutive” (“G”), occurrence in all isoforms with unchanged splice sites; “facultative” (“F”), occurrence in all isoforms with varying splice sites; and otherwise “alternate” (“A”). Different splice-site variants (5′, 3′, or both) are annotated with n, c, and b instances, tracked numerically for unique variants, and constituting Block-III in EUID. “N” in EUID descriptor (top row) is numeric character denoting occurrence count for respective block attributes, and “ISF” corresponds to isoform. (B) Exon annotation with each block is illustrated, focusing on transitions between coding, noncoding, and dual states from the reference state (coding) after it undergoes splice-site variations. Gray and light green colors indicate the exon's coding and noncoding scope. The GC and CGC are shown with the dashed line. An asterisk indicates that appropriate code will be inserted based on relevant information. (C) Variants exhibiting changes in Block-I scope owing to splice-site variation at respective exon positions are shown for genes POLI, SEC24B, THAP9, and ADORA1 (from left to right). Exon variants undergoing “dual” scope changes are specified with both “coding” and noncoding subtypes, connected by an arrow on right. The ID mentioned in brackets refers to Entrez gene identifiers. Corresponding abbreviations used in figure are expanded at their respective positions.











