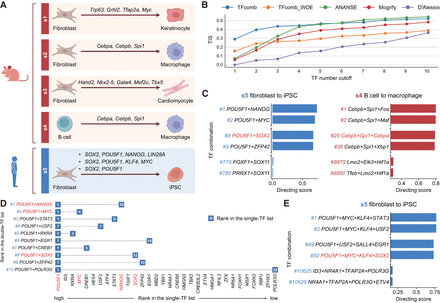
TFcomb identifies the TFs directing cell reprogramming across different scenarios. TFs here refer to genes that encode the corresponding TFs. (A) Five reprogramming scenarios (s1–s5) with experimentally validated reprogramming TF combinations. (B) The line plots show the comparison of different methods based on the TIS calculated between identified TFs and ground-truth reprogramming TFs. (C) Ranked TF combinations with TFcomb-predicted directing scores. The ground-truth combinations of case s4 (Cebpa, Cebpb, and Spi1) and s5 (POU5F1 and SOX2) are marked in red. (D) Comparison between the single-TF list and double-TF list of TFcomb in case s5. The single ground-truth reprogramming TFs in the rows and the double ground-truth reprogramming TFs in the columns are both marked in red. (E) Ranked TF combinations with TFcomb predicted directing scores. The Yamanaka factors are marked in red.











