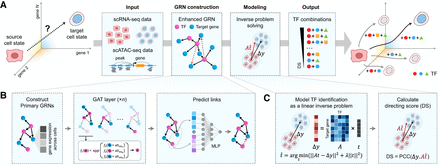
A graphical illustration of TFcomb. (A) Overview of TFcomb. TFcomb can identify the TFs and TF combinations that reprogram the source-cell state to the target-cell state. (B) TFcomb first constructs a primary GRN with scRNA data and scATAC data, and then it enhances the primary GRN with GAT. The normalized gene expression and the primary regulatory network comprise the input graph, and each node represents a gene. The whole model consists of the GAT encoder and the multilayer perceptron predictor. A multihead attention mechanism is applied in the GAT layer to stabilize the learning process. (C) The TF identification task is modeled as an inverse problem and solved with Tikhonov regularization. TFcomb uses the calculated expected alteration to get the directing score of each TF.











