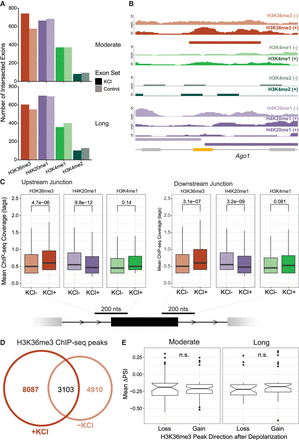
H3K36me3 dynamically marks a subset of alternative exons in response to KCl. (A) Number of KCl-dependent (saturated bar) and control (unsaturated bar) exons that overlap an intragenic H3K36me3 peak (Wang et al. 2015) across moderate and long KCl treatments. Exons and H3K36me3 peaks were considered overlapping with a >90% overlap. Equivalent analysis was performed for H4K20me1, H3K4me1, and H3K4me2. (B) Genome tracks of ChIP-seq coverage and peaks for H3K36me3, H3K4me1, H3K4me2, and H4K20me1 over a candidate KCl-dependent CE in Ago1. (C) Mean ChIP-seq coverage of H3K36me3, H4K20me1, and H3K4me1 at upstream and downstream exon–intron junctions (100 nt intron + 100 nt exon) of KCl-dependent alternative exons. A Mann–Whitney U test was used to test significance of mean ChIP coverage between treatments, and significance was considered at P < 0.05. (D) Euler plot of H3K36me3 peaks called between KCl and untreated primary cortical neurons. Peaks were considered overlapping with reciprocal overlap of >10%. (E) Mean ΔPSI of KCl-dependent exons under a changing H3K36me3 peak. Gain and loss were characterized as being unique to the +KCl (D, nonoverlapping left circle) or −KCl (D, nonoverlapping right circle) sets, respectively. An unpaired t-test was performed to assess difference in mean ΔPSI between exons under a KCl-dependent H3K36me3 peak that disappears (decreased) or appears (increased) after KCl stimulation.











