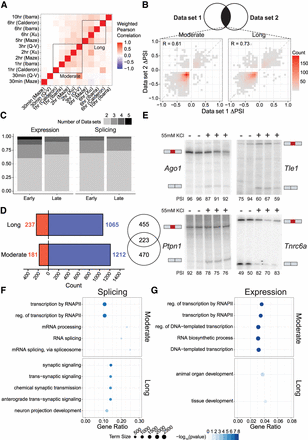
Integrative analysis defines activity-dependent exons of high confidence. (A) Heatmap of weighted Pearson's correlations between all data sets used in analysis (for description of calculation, see Methods). Data sets categorized as “moderate” and “long” after KCl stimulation are designated. Q-V is equivalent to Quesnel-Vallières et al. (2016). (B) Tile plot of ΔPSIs for shared events between pairwise data sets in the moderate (left) and long (right) classifications. Correlation is calculated using Pearson's R with significance considered P < 0.05. Plot space is organized into a 50 × 50 grid with greater color denoting a higher number of events mapping to a given tile. (C) Number of data sets in which a high-confidence event is significant for expression (left) and splicing (right). (D) Summary of all high-confidence events in the moderate and long alternative exon sets. Euler plot of gene-level overlaps between the two sets is shown at right. (E) 32P-labeled RT-PCR of four candidate high-confidence CEs in primary mouse cortical neurons stimulated with 55 mM KCl for 2 h. PSI values for each sample are quantified beneath the gel. (F,G) Enriched Gene Ontology terms for differential splicing (F) and differential expression (G) high-confidence events in the moderate and long sets. Terms are derived only from the “Biological Process” domain and are filtered to contain fewer than 2500 genes in each term. The top five terms by descending −log10(FDR) are shown for each condition, with the size of the shape corresponding to the number of genes annotated in a given GO term. Gene ratio is calculated as the proportion of genes identified in experimental samples over the total number of annotated genes in the GO term. Analysis of long high-confidence-set differential expression (G, lower) yielded only two significantly enriched GO terms.











