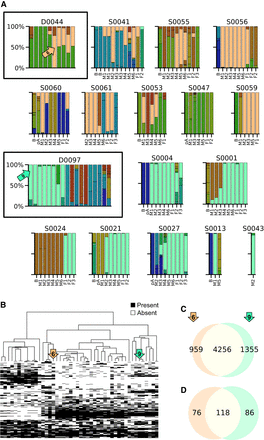
Different donors in a fecal microbiota transplant (FMT) trial (Smith et al. 2022b) have engrafting E. coli strains that differ in their functional potential. (A) E. coli strains found in repeated sampling of two independent donors’ fecal materials (boxed panels) and in the fecal time series of their respective recipients. Columns in each panel represent individual samples; colors represent E. coli strains inferred from StrainFacts; and the height of colored bars indicates strain abundance normalized to total E. coli abundance in the sample. For donors, samples are in arbitrary order. Recipient samples are ordered by collection day and include samples at baseline (B) collected before initial FMT treatment, samples collected before each of up to six maintenance FMT doses (labeled M1 to M6), and up to three follow-up samples (labeled F1 to F3). For a subset of recipients, samples were also collected after antibiotic treatment and before FMT (labeled pA for postantibiotics). For each donor, one strain (tan in D44, aqua in D97) showed a high rate of engraftment in recipients at follow-up. (B) Comparison of shell gene content between inferred strains from the FMT experiment (18 strains) and E. coli strains from the HMP2 (28). Heatmap indicates the presence and absence of genes (rows) across inferred strains (columns). Strains are ordered by UPGMA tree of estimated SNP genotype dissimilarity. Genes are filtered to only the 3134 genes in the shell pangenome fraction. Arrows (tan and aqua) highlight the high-engraftment strains from panel A. (C,D) Estimated gene content that is shared and distinct between the two high-engraftment strains. Venn diagrams depict the intersection of genes (C) and gene co-occurrence clusters (D).











