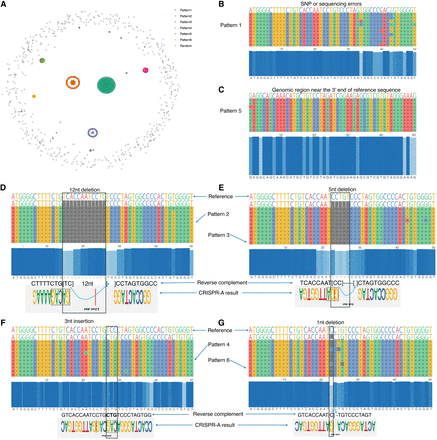
Gene editing patterns. (A) KMAP visualization of aligned DNA sequences. The six clusters are given by DBSCAN based on the 2D embeddings. (B–G) Sequence alignment of each pattern. Each row represents a DNA sequence. Ten sequences from each cluster are shown in the alignment to illustrate the corresponding pattern. The blue panel in the center shows the conservative nucleotide at each position. The top four gene editing patterns given by CRISPR-A are provided at the bottom panel for the last four patterns (D–G), above which are their reverse complements. Note that although the deletions in our result show 1–3 nt shifts compared with CRISPR-A's result, they are equivalent as the shifted nucleotides can be placed on either side of the deletion in the alignment. Pattern 1 is treated as SNPs or sequencing errors by CRISPR-A. Pattern 5 corresponds to a different genomic region.











