Table 1.
De novo motifs related to chromatin organization and regulation
| No. | Motifs | Best match | Functions | P-value |
|---|---|---|---|---|
| 1 |  |
CTCF | The functions of CTCF have been discussed earlier. | 1.00 × 10−16 |
| 2 | 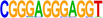 |
MAZ | MAZ works together with CTCF to control cohesin positioning and genome organization (Xiao et al. 2021). | 1.00 × 10−16 |
| 3 | 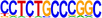 |
THAP1 | Cellular THAP proteins may function as zinc-dependent sequence-specific DNA-binding factors with roles in proliferation, apoptosis, cell cycle, chromosome segregation, chromatin modification, and transcriptional regulation (Clouaire et al. 2005). | 1.00 × 10−15 |
| 4 | 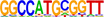 |
RUNX2 | Runx2/Cbfa1 diversely regulates gene transcription through chromatin remodeling and coregulatory protein interactions (Lian et al. 2003). Runx2 regulates chromatin accessibility to direct the osteoblast program at neonatal stages (Hojo et al. 2022). | 1.00 × 10−15 |
| 5 |  |
STAT5 | STAT3 and STAT5 have key interactions with chromatin remodeling factors such as DNA methyltransferases, histone modifiers, cofactors, corepressors, and other transcription factors (Wingelhofer et al. 2018). | 1.00 × 10−14 |
| 6 | 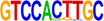 |
MYC | MYC proteins are required for the widespread maintenance of active chromatin (Knoepfler et al. 2006). | 1.00 × 10−10 |











