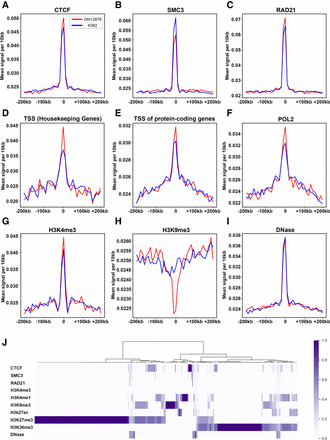
Hierarchical TAD boundaries are highly correlated with epigenetic factors. Enrichment of epigenetic factors near TAD boundaries identified by BINDER on GM12878 and K562 data, namely, CTCF (A), SMC3 (B), RAD21 (C), TSS of housekeeping genes (D), TSS of protein-coding genes (E), POL2 (F), H3K4me3 (G), H3K9me3 (H), and DNase (I), where the red line indicates GM12878, and the blue line indicates K562. (J) Hierarchical clustering of hierarchical TAD boundaries based on nine biological features and DNase-seq, where each value of a factor for each boundary is normalized to zero to one (for details, see the “Hierarchical Clustering for the Enrichment of Epigenetic Factors near TAD Boundaries” section in Supplemental Methods).











