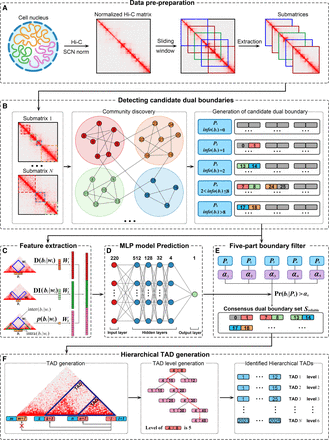
Workflow of BINDER pipeline. (A) BINDER uses a fixed-size sliding window to extract submatrices along the diagonal of a SCN-normalized Hi-C contact matrix. (B) The set of candidate dual boundaries Sdual is generated by applying Infomap to these submatrices (for the description of dual boundary, see Methods). (C) Three features are extracted for each dual boundary in Sdual, including the local interaction density, DI, and P-value of Wilcoxon rank-sum test, and then, they are concatenated to a 220-dimensional feature vector. (D) A trained neural network model with one input layer (containing 220 neurons), four hidden layers (containing 512, 128, 32, and four neurons, respectively), and one output layer (outputting predicted reliability score for each boundary). (E) The set of consensus dual boundary Sreliable is generated by both the reliability scores predicted by the MLP model and the occurrence counts during the community discovery part by Infomap reaching a consensus on boundaries from Sdual. (F) Hierarchical TADs and their hierarchies are finally obtained by the left-neighbor extension and hierarchy generation methods.











