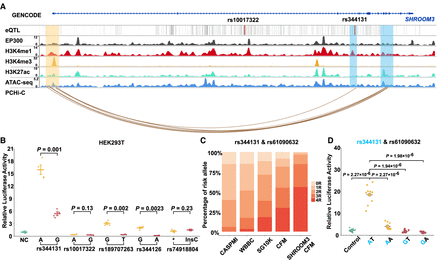
In vitro analysis of eQTL effects on enhancer activity. (A) Epigenetic landscape at SHROOM3 in human neural crest cells (hCNCCs). This panel presents an integrated epigenomic analysis, combining in vitro PCHi-C, ATAC-seq, and ChIP-seq data from hCNCCs (Rada-Iglesias et al. 2012; Xu et al. 2024). It displays the epigenetic markers H3K4me1, H3K4me3, and H3K27ac in hCNCCs. The promoter is highlighted with light orange shading. Enhancers are highlighted with blue shading, with eQTLs within these enhancers marked by black lines (darkness represents the significance level). In the PCHi-C section, differential chromatin interactions connect enhancers to the SHROOM3 promoter, with line colors indicating the strength of interaction. (B) Luciferase assays to assess the effects of eQTLs on SHROOM3 enhancers, including an empty vector as a control. Data from three independent experiments, each comprising three technical replicates, are depicted. (C) Enrichment analysis of risk allele combinations for eQTLs rs344131 and rs61090632 in CFM patients compared with control populations. Significant enrichment is observed in both the entire CFM cohort (P < 0.008884) and the SHROOM3 CFM subset (P < 1.71 × 10−24) using chi-square tests. Control populations: (CASPMI) Chinese Academy of Sciences Precision Medicine Initiative, (WBBC) Westlake BioBank for Chinese, and (SG10K) Singapore 10K Genome Project. Patient cohorts: CFM (n = 2009) and SHROOM3 CFM (n = 361). (D) Luciferase assay results demonstrating the combined effect of alleles rs344131 and rs61090632 in HEK-293T cells. P-values are calculated using the Student's t-test.











