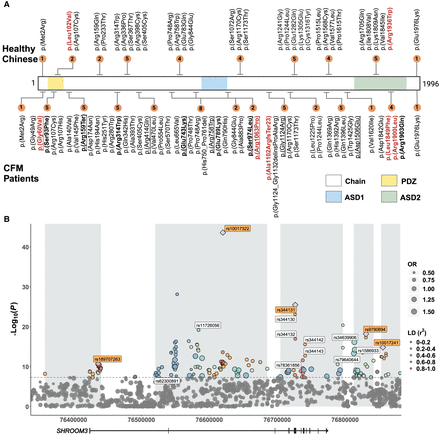
Distribution of putative deleterious variants and common SNP associations in SHROOM3 among CFM patients and healthy Chinese. (A) The schematic delineates the PDZ, ASD1, and ASD2 domains of the SHROOM3 protein, color-coded in yellow, blue, and green, respectively. The upper panel enumerates the deleterious variants identified in healthy Chinese individuals, and the lower panel details those found in CFM patients, with variants from European/Hispanic/Australasian ancestry underlined. The count of deleterious variants within specific regions is indicated by numbered brown circles. Variants emphasized in red denote those predicted as pathogenic by the ESM1b and AlphaMissense algorithm. Likely pathogenic variants determined using combination patterns are marked by bold in CFM patients. (B) Manhattan plot of CFM-associated variants in SHROOM3. This plot showcases variants with a minor allele frequency above 0.05. The size of each dot indicates the odds ratio of the corresponding variant. The plot demarcates five linkage disequilibrium (LD) blocks within SHROOM3 in gray, with the color-coding reflecting r² values, signifying the correlation between the lead SNP and other variants within each LD block. Lead SNPs are marked with purple rhombus, and their rsID is shown with an orange background. eQTL sites are identified with rsIDs with a white background. (OR) Odds ratio.











