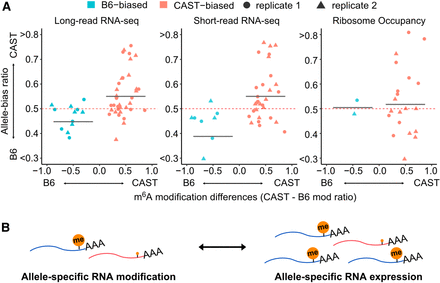
Effects of ASM on allele-specific RNA expression and ribosome occupancy. (A) Allele bias ratio of genes containing ASM sites (sky blue for B6 biased; pink for CAST biased). Different shapes represent the replicates (circle for replicate 1, triangle for replicate 2) The y-axis displays the allele bias ratio obtained from long-read (left), short-read (middle) sequencing, and ribosome profiling (right). The x-axis shows the difference in m6A modification ratios between the two alleles (CAST–B6). The red dashed horizontal bar indicates an allele-bias ratio 0.5, an allele-bias cutoff point. The gray bar represents the mean allele bias ratio for genes with B6 or CAST-biased ASM sites. (B) Model for regulation of ASM and ASE. The two-sided arrow in the model reflects the possibility of bidirectional influence, rather than suggesting unidirectional causality. Differential transcription driven by allele-specific epigenetic contexts may regulate ASM, indicating that both phenomena could share a common underlying mechanism, without one necessarily being the direct cause of the other.











