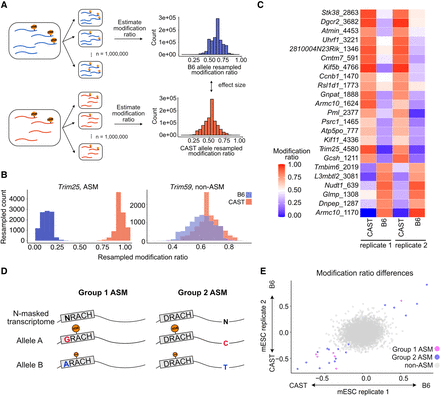
Identification and classification of ASM sites. (A) Schematic of statistical procedure for ASM detection (Methods). Reads overlapping the site under consideration were resampled, and the modification ratio was estimated in each bootstrap sample. A statistically significant ASM site was defined as an adjusted harmonic P-value (FDR < 0.1; Methods). (B) An ASM site within Trim25, exhibits distinct modification ratio samples. Conversely, a non-ASM site within Trim59 displays substantial overlap in modification ratios between bootstrap sampling distributions. (C) Modification ratios of each allele across mESC wild-type replicates. The y-axis displays the name of the gene and m6A position in the transcript. (D,E) ASM sites were classified into two groups. Group 1 is defined by genetic variants within the DRACH motif, and Group 2 is characterized by variants adjacent to or distal from the DRACH motif (D). The modification differences of the defined ASM were represented by color according to their classification (Group 1 in magenta, Group 2 in blue, and non-ASM in gray). Each axis is the modification ratio, where negative values denote CAST allele bias and positive values indicate B6 allele bias in m6A modification (E).











