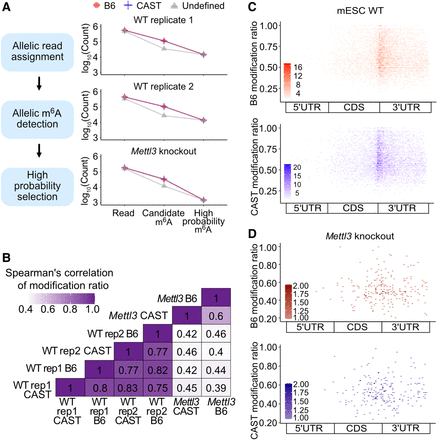
Comparative analysis of allelic modifications in wild-type (WT) and Mettl3 knockout mESCs. (A) Allelic impartiality while allelic read assignment and m6A detection. The left panels outline the schematic of the data procedural steps for ASMs analysis. The right panels display the counts for allelic reads, candidate m6A sites, and high-probability m6A sites selected through our criteria (red circles, B6; blue plusses, CAST; and gray triangles, undefined group). The left top two plots show the counts from mESC WT replicates and the bottom plot exhibits the numbers from mESC Mettl3 knockout. (B) Spearman's correlation of modification ratio between alleles from WT and Mettl3 knockout cells (rep1: mESC WT replicate 1; rep2: mESC WT replicate 2; Mettl3: Mettl3 knockout). (C,D) The distribution of sites with high probability of m6A modification (prob > 0.85) is displayed in a metagene plot by calculating the relative positions of these sites within gene regions. The color scale represents the number of m6A sites with the given modification ratio inferred from reads assigned to either of the two alleles in mESC WT (C) or Mettl3 knockout (D) cells (red, B6 allele; blue, CAST allele).











