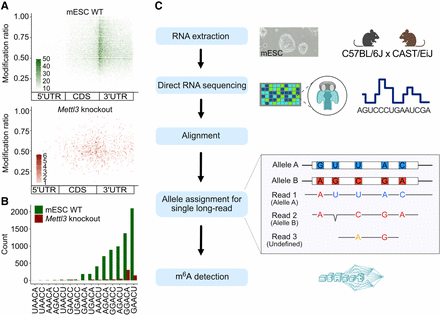
Allelic read assignment and m6A modification analysis using ONT DRS in hybrid mESCs. (A) m6A modification ratio and locations detected from m6Anet using all reads (top, green, WT; bottom, red, Mettl3 knockout). The relative m6A locations within the transcript body were determined. It presents modification ratios after high-probability selection (>0.85). The color darkness represents the counts of the ratio on the position. (B) Comparison of the frequencies of instances of DRACH motif sequences (green, WT; red, Mettl3 knockout). (C) Schematic overview of the strategy used for allelic long-read assignment for ASM analysis. Total RNA from hybrid mESC (C57BL/6J × CAST/EiJ) underwent DRS. To avoid reference bias, we used an N-masked transcriptome for alignment. Reads were then allocated to each allele. This process classified reads into Allele A (B6), Allele B (CAST), and undefined categories, enabling m6A detection within each group individually.











