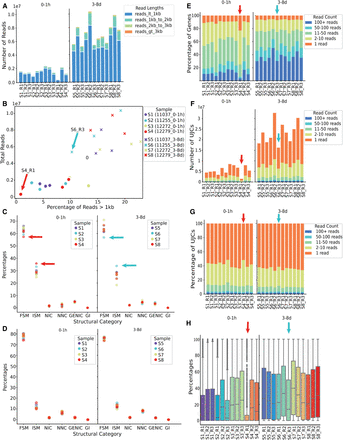
SQANTI-reads analysis of Drosophila samples. (A) Number of mapped reads by experimental group labeled with read length. (B) Percentage of mapped reads >1 kb versus percentage of reads that are FSM. Dots represent the early stage (0–1 h after eclosion) and crosses indicate the adult stage (3–8 days old). The four genotypes are indicated with four different colors. (C) Percentage of reads mapping to genes in each SQANTI3 structural category (D) Percentage of reads mapping to genes in each SQANTI3-QC structural category for reads >1 kb (E) Number of genes detected with breakdown by the number of reads mapped to each gene. (F) Number of UJCs detected with breakdown by the number of reads associated with each UJC. (G) Percentage of UJCs detected with breakdown by the number of reads associated with each UJC. (H) Distribution of the percentage of FSM reads by gene across samples.











