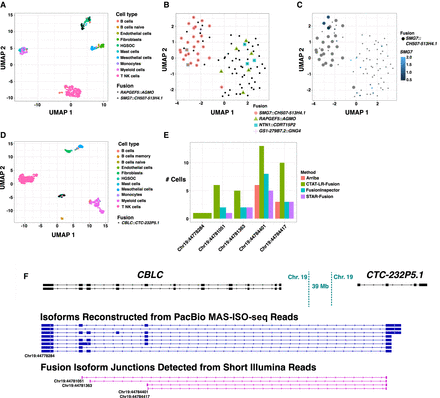
Fusion expression intratumor heterogeneity observed in high-grade serous ovarian cancer cells. (A) UMAP embedding of all cells from HGSOC patient-1, colored by cell type. Fusion RAPGEF5::AGMO and SMG7::CH507-513H4.1 are expressed in two different HGSOC cell clusters. Fusion-containing cells are shown as black points with shape according to fusion gene pairs. (B) UMAP embedding of HGSOC cells from HGSOC patient-1, colored by fusions expressed. RAPGEF5::AGMO is expressed exclusively in the right cluster. SMG7::CH507-513H4.1 and GS1-279B7.2::GNG4 fusions coexpress and are expressed almost exclusively in the left cluster. The two NTN1::CDRT15P2 fusion-expressing cells in the left cluster coexpress the SMG7::CH507-513H4.1 fusion. (C) Tumor cells are colored according to SMG7 expression and cells identified with fusion SMG7::CH507-513H4.1 are shown as large dots. Several cells are found to robustly express SMG7 that do not have evidence of expressing the fusion transcript. (D) UMAP embedding for all cells from HGSOC patient-3, colored by cell type. Cells expressing fusion CBLC::CTC-232P5.1 are shown as black dots. (E) Numbers of cells found to express each of the five alternative splicing fusion breakpoints for fusion CBLC::CTC-232P5.1 according to long- or short-read support and corresponding search method, with CBLC fusion isoform breakpoints labeled according to GRCh38 coordinates. (F) Fusion-supporting transcript isoform structures based on long-read (center) or short-read (bottom) sequences in the context of the FusionInspector modeled gene fusion contig. GENCODE v22 reference isoform transcript structures for CBLC and CTC-232P5.1 genes are shown at top. CBLC fusion isoform breakpoints corresponding to (E) are labeled accordingly.











