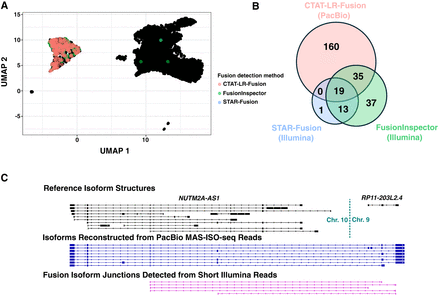
Detection of Fusion NUTM2A-AS1::RP11-203L2.4 in a T cell infiltrated melanoma tumor sample. MAS-Iso-Seq and matched Illumina RNA-seq data from a melanoma tumor sample M132TS 10× single-cell library (published in Al'Khafaji et al. 2023) were examined for fusion transcripts using CTAT-LR-Fusion for PacBio long reads and STAR-Fusion, FusionInspector, and Arriba for Illumina short reads. (A) UMAP for melanoma sample M132TS single cells. Individual cells are shown as (mostly overlapping) black dots. Cells identified with the NUTM2A-AS1::RP11-203L2.4 fusion transcript are colored according to the detection method, predominantly labeling the cluster of malignant cells. (B) Venn diagram indicating the numbers of fusion-containing cells according to detection methods. Fusion NUTM2A-AS1::RP11-203L2.4 was not reported by Arriba. (C) Fusion-supporting transcript isoform structures based on long (center) or short (bottom) read sequences in the context of the FusionInspector modeled gene fusion contig. GENCODE v22 reference isoform transcript structures for NUTM2A-AS1 and RP11-203L2.4 genes are shown at top.











