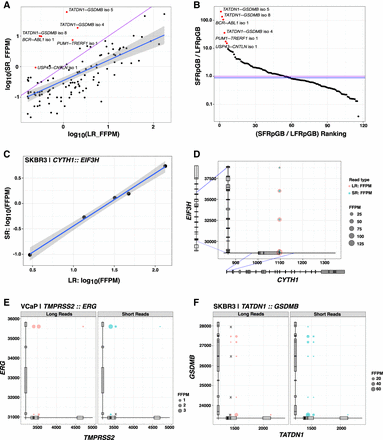
Comparison of long (MAS-ISO-seq) versus short read (TruSeq Illumina) support for fusion isoforms. (A) Comparison of fusion isoform read evidence quantification according to CTAT-LR-Fusion (MAS-ISO-seq long reads) and FusionInspector (TruSeq Illumina short reads), with read support normalized for sequencing depth as fusion fragments or reads per total million reads (FFPM). (B) Ranking of fusions according to the ratio of Illumina short fusion reads per gigabases sequenced (SFRpGB) to PacBio long fusion reads per gigabases sequenced (LFRpGB). Identity line is shown in purple, and the median ratio is shown in blue. (C,D) Five fusion isoforms observed for the fusion gene CYTH1::EIF3H of cell line SKBR3 are (C) observed with highly correlated expression measurements as estimated from long and short RNA-seq reads and (D) shown according to fusion transcript breakpoints. (E,F) Read alignment evidence quantified for fusion isoform breakpoints according to long-read (left, red) or short-read (right, blue) RNA-seq: (E) TMPRSS2–ERG fusion in VCaP, (F) TATDN1–GSDMB fusion in SKBR3. “X” indicates missing read support for the corresponding isoform in the alternative sequencing type.











