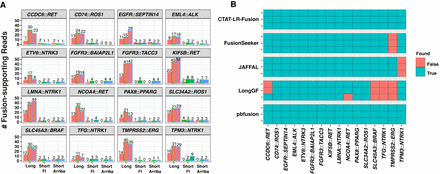
Fusion transcript detection applied to SeraCare v4 Fusion Reference Control sample. (A) Quantities of PacBio long reads and TruSeq Illumina short reads identified as evidence for each of the 16 control fusions as ascertained by CTAT-LR-Fusion (long reads), FusionInspector (short reads), and Arriba (short reads) across each sample replicate. PacBio replicate reads were down-sampled to match the number of sequenced bases from the respective Illumina replicate samples. (B) Binary heatmap for the identification of the 16 control fusion pairs by different fusion detection software according to each of the three replicates of long-read sequences, using all (not down-sampled) sequenced reads. PacBio replicates are ordered (A) left to right or (B) top to bottom as MAS-ISO-seq monomer (replicate 1), and MAS-ISO-seq 8mer-concatamer sequenced replicates 2 and 3. Counts of sequenced reads are provided in Supplemental Table S1.











