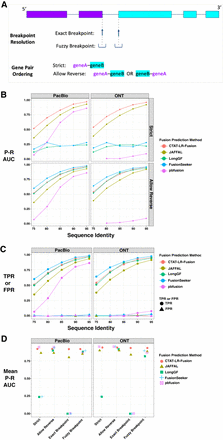
Accuracy for fusion transcript detection using simulated long reads. (A) Scheme for criteria in benchmarking fusion detection. (B) Accuracy is reported as P–R AUC using simulated PacBio and ONT long reads with moderate to high error rates (test data derived from Davidson et al. 2022), requiring proper fusion gene pair ordering (Strict) or allowing for either gene pair ordering (Allow Reverse). (C) True positive rate (TPR) and false positive rate (FPR) were observed for each method and data set at corresponding peak F1 scores, allowing for either fusion gene pair ordering (Allow Reverse). (D) Accuracy using PBSIM3 simulated PacBio HiFi or ONT R10.4.1 isoform reads at 5× coverage additionally focused on breakpoint resolution, with mean of P–R AUC values across five samples of 500 different target fusions each. Similar findings resulted at 50× coverage (Supplemental File 2).











