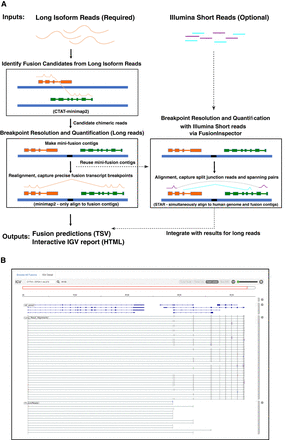
CTAT-LR-Fusion workflow and output. (A) CTAT-LR-Fusion workflow: Long isoform read sequences and optional matched short RNA-seq reads are provided as input. Long reads are aligned to the reference genome using ctat-minimap2, a slightly modified minimap2 tool that yields alignments only for reads that map to multiple genomic loci. Reads identified as candidate chimeric reads are further assessed as fusion evidence by modeling fusion contigs containing the candidate fusion genes in their proper order and orientation followed by realignment of candidate chimeric reads to the fusion contigs using regular minimap2. Fusion transcript breakpoints and fusion read support are quantified in the context of the fusion contigs. If matched short reads are available, FusionInspector (included with CTAT-LR-Fusion) uses STAR to simultaneously align the short reads to the fusion contigs and the human reference genome, yielding fusion evidence quantification based on short-read RNA-seq. The FusionInspector findings based on short-read RNA-seq are then integrated into the final fusion report generated by CTAT-LR-Fusion, specifying the fusion evidence support for fusion transcript isoforms by long and/or short reads according to breakpoint. Outputs include tab-delimited reports describing the fusion alignment evidence and quantifications and an interactive IGV-report in HTML format for navigating the fusion alignment evidence. (B) Example IGV-report highlighting evidence for fusion CYTH1::EIF3H in cell line SKBR3. The IGV-report visualization provides interactive analysis of reference annotation gene structures (top) and long isoform read alignment evidence for predicted fusion transcripts (center) and includes alignments for matched Illumina short reads where available (bottom). In this example, multiple alternatively spliced fusion isoforms are evident from both long and short reads, and most long reads appear to yield full-length fusion transcript isoforms, whereas the short reads primarily yield fusion isoform breakpoint positions.











