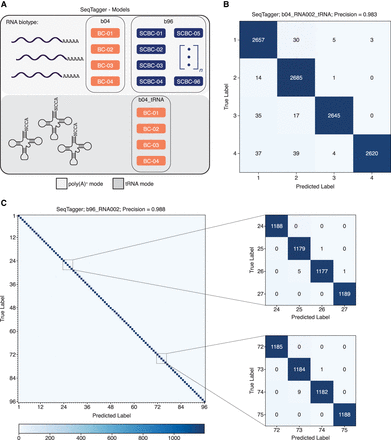
SeqTagger can be expanded to work on RNA–DNA hybrids (Nano-tRNAseq libraries) and with larger sets of barcodes. (A) Schematic overview of the demultiplexing models supported by SeqTagger. A four barcode (b04) and 96 barcode (b96) model are available. Additionally, a four barcode demuxing model is available for custom Nano-tRNAseq libraries (b04_tRNA). (B) Confusion matrix for the four barcode tRNA model (b04_tRNA) generated on Nano-tRNAseq validation data. Recorded precision is indicated on top. (C) Confusion matrix of the 96 barcode mRNA model (b96_RNA002) generated on the validation data. Recorded precision is indicated on top. Zoomed panels of the 96 × 96 confusion matrix for eight barcodes are shown on the right.











