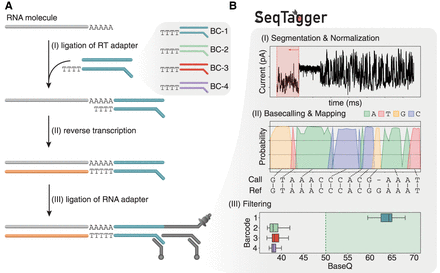
Schematic overview of the DRS and demultiplexing workflow. (A) Overview of the barcoded DRS workflow in which the standard RTA is replaced with a barcode-containing adapter. Following adapter ligation and reverse transcription, the RNA ligation adapter (RLA), containing the helicase enzyme, is ligated, making the library sequencing-ready. (B) Overview of the demultiplexing workflow performed by SeqTagger. The algorithm first segments the raw current intensity signal by identifying the poly(A)-tail signal to extract the barcode-containing reverse transcription adapter (RTA). Following signal normalization, the DNA sequence is basecalled and aligned to a set of reference barcodes. Finally, a filtering step is applied based on the median base quality (baseQ) to remove misassigned barcode sequences.











