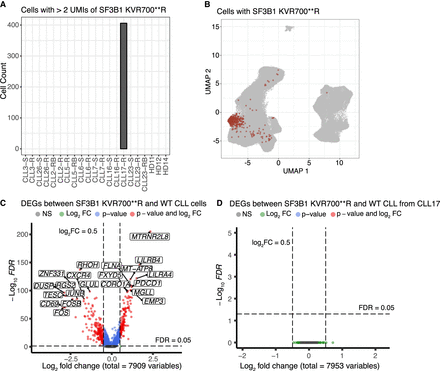
The SF3B1 KVR700**R alteration is detected by scRaCH-seq. (A) Bar plot showing the cells with SF3B1 6 bp deletion (Chr 2:197,402,104:197,402,109) across all samples. A criterion of >2 of SF3B1 with 6 bp del transcripts (UMIs) per cell was employed. (B) UMAP projection of cells carrying >2 SF3B1 KVR700**R altered transcripts (red). (C) Volcano plot showing the DEGs (FDR < 0.05) between SF3B1 KVR700**R mutant and wild-type CLL cells from all venetoclax-relapsed CLL samples. (D) Volcano plot showing the DEGs (FDR < 0.05) between SF3B1 KVR700**R mutant and wild-type CLL cells from CLL17.











