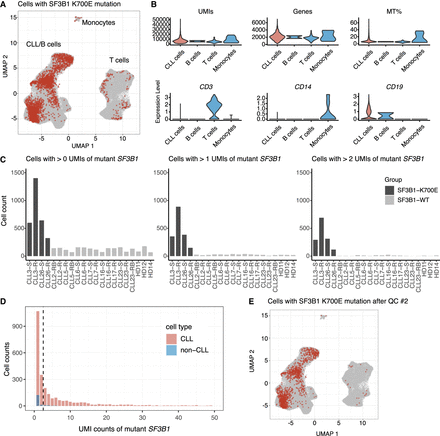
The SF3B1 K700E mutation is detected by scRaCH-seq. (A) UMAP projection of cells carrying the SF3B1 K700E mutation (red). (B) Violin plot showing the UMI counts, gene counts (Features), expression of mitochondria genes (MT%), expression of CD3 (T cell marker), CD14 (monocyte marker), and CD19 (B cell marker) per cell type cluster. (C) Bar plot showing the abundance of mutant transcripts. Left plot showing cells per sample with >0 transcripts (UMI) of mutant SF3B1 K700E. Middle plot showing cells per sample with >1 transcripts (UMIs) of SF3B1 mutation. Right plot showing cells per sample with >2 transcripts (UMIs) of SF3B1 mutation. Samples with confirmed SF3B1 K700E mutation by WES are highlighted in dark gray. (D) Bar plot showing the distribution of the abundance of SF3B1 K700E mutation detected in CLL (pink) and non-CLL (blue) cells. The vertical dashed line indicates the criterion of >2 SF3B1 mutation transcripts. (E) UMAP projection of cells carrying >2 SF3B1 K700E mutation transcripts (red).











