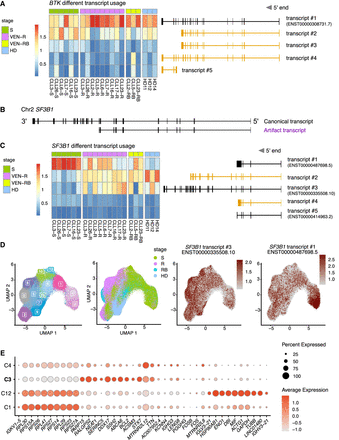
scRaCH-seq reveals different SF3B1 isoform usage by CLL cells from patients undergoing different treatments. (A) Heatmap showing BTK isoform usage (rows) per sample (columns) in the B/CLL cells. The top 5 BTK transcripts are illustrated on the right with novel transcripts highlighted in yellow. Samples are grouped in screening (S; green), venetoclax-relapsed (VEN-R/R; pink), venetoclax-relapsed and subsequently on BTKi (VEN-RB/RB; bright yellow), and HD (turquoise). (B) Illustration of the dominant SF3B1 transcript identified and characterized by the absence of the last 14 exons in the 3′ end (highlighted in purple). (C) Heatmap showing SF3B1 isoform usage (rows) per sample (columns) in the B/CLL cells. The top 5 SF3B1 transcripts are illustrated on the right with novel transcripts highlighted in yellow. (D) UMAP projection of CLL/B cells from CLL patients or HDs and clustering based on short-read gene expression. The samples from CLL patients at screening (S; green), venetoclax-relapsed (R; pink), venetoclax-relapsed and subsequently on BTKi (RB; bright yellow) and HDs (turquoise) are highlighted in the UMAP. The expression of the SF3B1 transcripts #1 and #3 is overlaid as red dots in the UMAP (B). (E) Dot plot showing marker genes that distinguish the CLL cells present in the four clusters shared by the screening samples (C1, C3, C4, and C12 in C). Cluster 3 with SF3B1 transcript #1 usage is indicated in bold.











