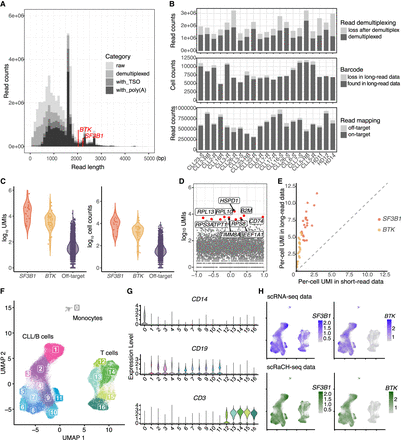
scRaCH-seq is efficient in capturing enriched genes of interest. (A) A graph showing the losses in read counts due to demultiplexing and integrity checks, depicting the distribution of read lengths ranging from 0 to 5000 bp. The canonical isoform of BTK (2027 bp) and SF3B1 (2225 bp) are marked on the plot. The four shades of gray, ranging from light to dark, correspond to raw reads, demultiplexed reads, reads with TSO, and reads with also a poly(A) end. (B) Bar plots showing the loss of reads after demultiplexing (top panel), barcodes detected in long-read sequencing data (middle panel), and counts of off-target and on-target reads for each sample (bottom panel). (C) Violin plot showing the log10 UMI counts of collapsed BTK (red), SF3B1 (orange), and off-target transcripts (purple) based on isoform detected by scRaCH-seq (left panel). Violin plot showing the log10 counts of cells possessing collapsed transcripts of BTK (red), SF3B1 (orange), and off-target genes (purple) (right panel). (D) Dot plot showing the log10 counts of off-target genes with the top 10 off-target transcripts specifically marked. (E) Dot plot showing the per-cell UMIs of SF3B1 (red) and BTK (orange), detected by scRNA-seq (x-axis) and scRaCH-seq (y-axis) for all samples. (F) Uniform Manifold Approximation and Projection (UMAP) projection of peripheral blood mononuclear cells from CLL patients or healthy donors and clustering based on short-read gene expression. (G) Violin plot showing the expression of CD14 (monocyte marker), CD19 (B/CLL cell marker), and CD3 (T cell marker) per cell cluster (F). (H) UMAP projection of SF3B1 (left) and BTK (right) gene-level expression detected by scRNA-seq (purple) or scRaCH-seq (green).











