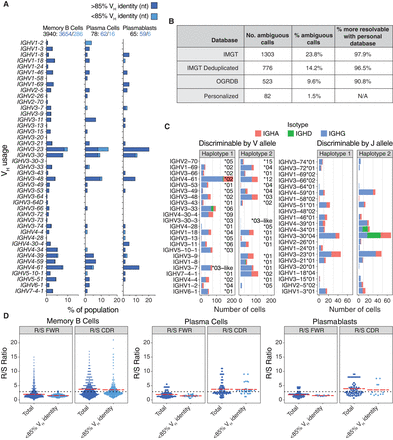
Donor-specific V gene usage and mutation rates. (A) Overall IGHV gene usage frequency for all complete and productive heavy chain consensus sequences with an in-frame CDR3. Cells with a low V gene identity, high mutation rates (≥15%) are stratified (light blue), as they are sometimes filtered out by programs such as Hi-V-QUEST (IMGT). (B) Statistics for ambiguous IGH and IGL V gene calls when using IgBLAST with various databases. (C) V gene usage for cells expressing one of the heterozygous V alleles (left), or that expressed a homozygous V allele but were haplotype-discriminable by a heterozygous J allele (right). (D) Ratio of replacement to silent mutations (R/S) in framework regions (FWRs) versus CDRs. The dotted line represents theoretical R/S ratios for unselected CDRs at R/S = 2.9. The red lines are the mean R/S values. The elevated replacement mutations in the CDRs suggest cells that have undergone antigen selection.











