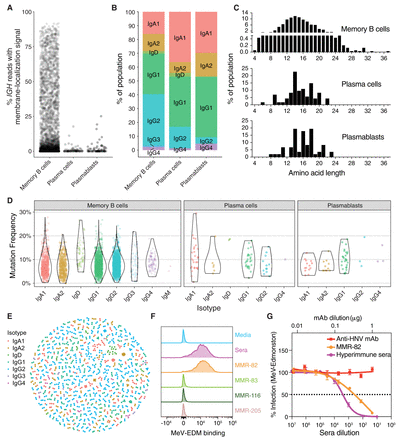
Characterizing single-cell IG sequences and antibody synthesis. (A) The percentage of IGH reads in each cell possessing the M exons indicative of a membrane-bound antibody, stratified by cell type. (B) Proportions of each isotype identified from consensus IGH sequences, stratified by cell type. (C) CDR3 amino acid length distributions for all productive IGH consensus sequences, stratified by cell type. CDR3 lengths are enumerated between the junction anchor residues: Cys 104 and Try 118 (IMGT numbering) for each sequence. (D) Mutation frequency compared to the germline sequences in the FWR1–4 and CDR1–3 regions for each cell type and isotype. (E) Dandelion plot of sequences clustered within each nonsingleton clone by VDJ amino acid sequence similarity. (F) Putative positive recombinantly expressed antibodies sequenced from the ASC gate were screened by flow cytometry for binding to MeV surface expressed proteins (F or RBP/H) on virus infected Raji-DC-SIGN B cells (multiplicity of infection [MOI] 0.01). MMR-vaccinated, hyper-immune sera from the donor at the time of cell collection was used as a positive control. (G) Neutralization assay in Vero cells for MMR-82 (MeV+) and hyper-immune sera against a GFP-expressing MeV-Edmonston recombinant virus (1000 IU/96-well). Data are presented as percent infection (mean ± SD) compared to infection in media alone (set as 100%). Experiments were performed in triplicate. Anti-HNV monoclonal antibody (mAb) is an irrelevant IgG1 mAb against an unrelated virus and serves a negative control.











