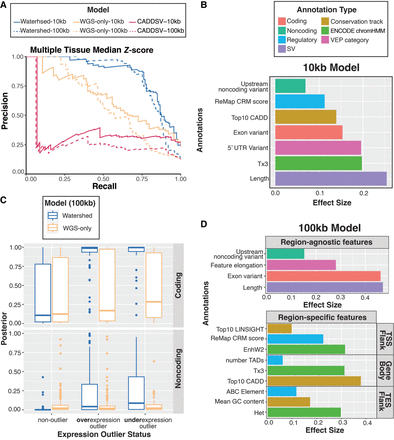
Watershed-SV improves the prioritization of rare SVs in healthy and muscular dystrophy cohort. (A) Precision-recall curves (PRC) of benchmark using held-out N2 pairs; We ran multitissue Watershed-SV using both 10 kb (solid) and 100 kb (dashed) distance limit as well as WGS-only model and CADD-SV with the same setup. (B) Top positive genomic annotation effect sizes (β) for seven major categories of the 10 kb multitissue Watershed-SV model. (C) Using a Z-score threshold of −3 and 3, we stratified 100 kb multitissue Watershed-SV model prediction on CMG muscular disorder data set posterior probabilities by under-, over-, and nonoutliers (column), and then by coding versus noncoding variants (row); each dot represent an gene-SV pair. (D) Top positive genomic annotation effect sizes for 100 kb multitissue Watershed-SV model. Seven annotation categories are grouped into region-specific (TSS/upstream Flank, Gene Body, TES/downstream Flank) and region-agnostic features. Region-specific features are separately aggregated for each SV, then collapsed to each gene by regions.











