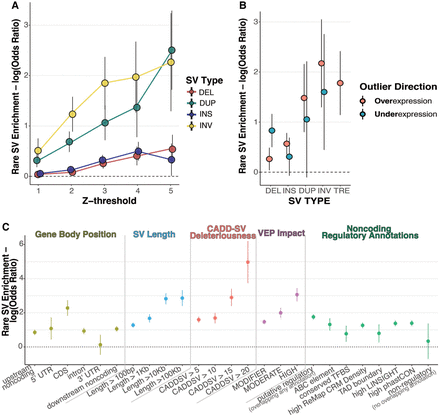
Rare long-read-discovered SVs are strongly enriched proximal to gene expression outliers (A) enrichment of rare SVs, stratified by type, within 10 kb of an expression outlier gene given the specified absolute Z-score threshold. Estimate of log odds ratio plotted with error bars representing standard errors of the estimate. (B) Directional enrichment for rare SVs within 10 kb of either over (Z > 4) or under (Z < −4) expression outliers. TRE-underexpression enrichment not plotted due to an insufficient number of rare TREs near underexpression outliers. (C) Enrichment of rare SVs, across all SV types, within 100 kb of expression outliers, stratified by genome and variant annotation categories. Gene body position displays enrichment of VEP annotated categories for SV location relative to the gene body of the expressed gene. If an SV overlaps multiple categories, it is assigned to the one with highest priority given the following ordering: CDS, 5′ UTR, 3′ UTR, intron, upstream noncoding, downstream noncoding. SV length and CADDSV deleteriousness display enrichment of rare SVs with length and CADDSV score respectively above the specified threshold. VEP impact displays enrichment of rare SVs with the given VEP impact category, where HIGH represents predicted loss-of-function variants. Finally, we display enrichment of SVs that overlap with noncoding regulatory annotations, including if it overlaps an ABC regulatory element linked to the expressed gene, a conserved transcription factor binding site (TFBS), a high density of ChIP-seq peaks defining conserved regulatory modules (CRM) from ReMap, a TAD boundary detected in multiple cell types, highly constrained LINSIGHT SNVs, or a highly conserved region by phastCons. We also display enrichments for SVs that overlapped any one of these annotations (putative regulatory SVs) and for SVs that do not overlap with any of these annotations (putative nonregulatory).











