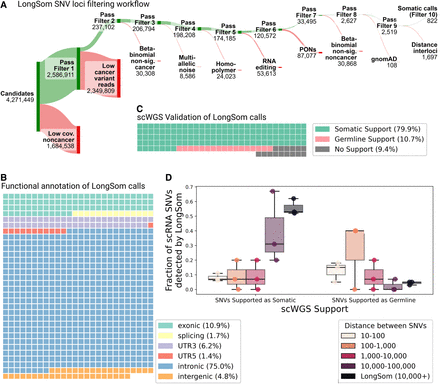
Validation of LongSom somatic calls using scRNA and scWGS data. (A) LongSom SNV filtering workflow, indicating the number of loci passing each of the 10 filtering steps (top, green) or being filtered (bottom, red), starting from all loci with at least one mutated read and five reads coverage in cancer cells. (B) Waffle plot representing each of the 822 somatic SNVs detected by LongSom, colored by their RefSeq functional annotation. (C) Waffle plot of all loci called by LongSom with sufficient power in scWGS for validation (Methods), colored by their status in scWGS. (D) Boxplot of the fraction of all loci called after filtering step 9 that is supported by scWGS data as either somatic or germline, colored by the distance from the closest mapping SNV also detected. LongSom calls represent all 822 calls after filtering step 10 (SNVs not within 10,000 bp or less from each other). Each point is a patient.











