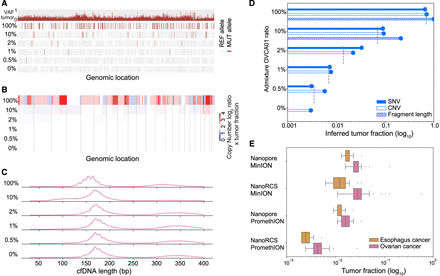
MRD detection using NanoRCS. (A–D) Admixture experiment with different percentages of OVCA01 versus HC02 cfDNA (100%, 10%, 2%, 1%, and 0.5%, 0% mixtures). (A) SNV observations for all dilutions. The top row shows the VAF of detected mutations in the tumor biopsy and the following five rows represent the MUT or REF allele observations in the different admixtures. (B) CNAs for all dilutions. Red indicates copy number gain and blue indicates copy number loss. Color intensity indicates the CNA multiplied by the TF in cfDNA. (C) cfDNA fragmentation length profiles for all dilutions. The profiles display the distribution of cfDNA fragment sizes ranging from 30 to 700 bp. (D) Inferred TF of each of the three NanoRCS modalities is shown in log-scaled bars. SNV (solid), CNA (nofill), and fragmentation length (striped) are compared to theoretical expected TFs (gray dashed lines). (E) Simulation results to assess the lowest TFs detectable for different platform throughputs (MinION vs. PromethION) and with or without consensus calling for Esophagus and OVCA. Simulation characteristics were obtained from PCAWG (see Methods).











